iGEM stands for the International Genetically Engineered Machine. It is the largest and most prestigious synthetic biology competition in the world. Founded by MIT, iGEM combines biology, engineering, entrepreneurship, etc., fostering teamwork, communication and passion.
At the 2025 iGEM Finals, which concluded just two weeks ago, the SCIE iGEM team: GreatBay-SCIE won the highest awards—the Grand Prize, along with the Gold Medal, Best Integrated Human Practices, Best Model, Best Education, Best Wiki, Best Village Prize, and a nomination for Best Presentation.
Last year, our iGEM school team GreatBay-SCIE 2024 achieved first runner up globally, along with multiple individual awards and nominations. We once thought that last year’s performance would be our limit, and the question, “Can we do better than last year?” often lingered jokingly in our conversations.
When we discovered during the summer vacation that the DNS reagent we had been using had expired for a long time, we even believed that the gold medal was out of our reach. Out of our expectation, this year, we made school history, becoming the first official school team to stand on the podium as the top in the world, while also securing nearly all available individual special prizes.
“Gooood afternoon! We are —— GREATBAY-SCIE!”
On the stage for the Finalist presentation, our voices once again resonated through the Grand Jamboree hall. The only difference was that when the finalist rankings were announced, we stood on the right-hand side of the stage — that’s right, the position for the champion.

Each team could compete for a maximum of three special prizes, yet we managed to achieve all three available special prizes that we’ve chosen, in addition to other general awards, such as Best Wiki and Best Village Prize.

Our story began in early March. When we noticed that one of our team members was troubled by the musty smell at home, we set our goal for the year: mold removal.
After initial research, we discovered that mold issues are widespread, yet public awareness of their actual hazards remains strikingly low. Moreover, most commercial mold removers rely on strong oxidizing agents, which often produce pungent odors and pose additional health risks. This led us to conceptualize our product: ArMOLDgeddon.
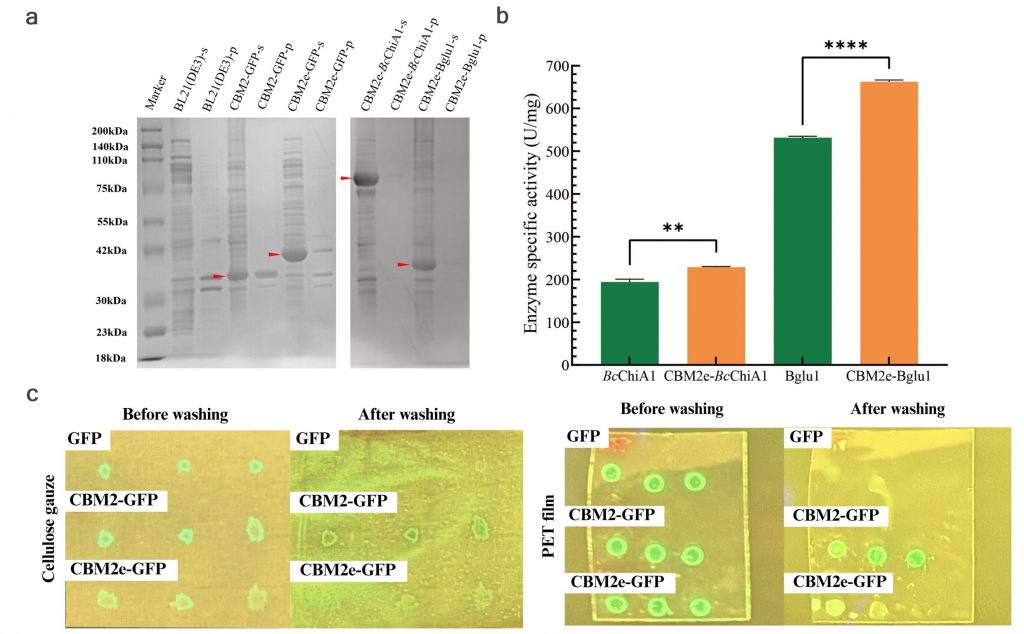
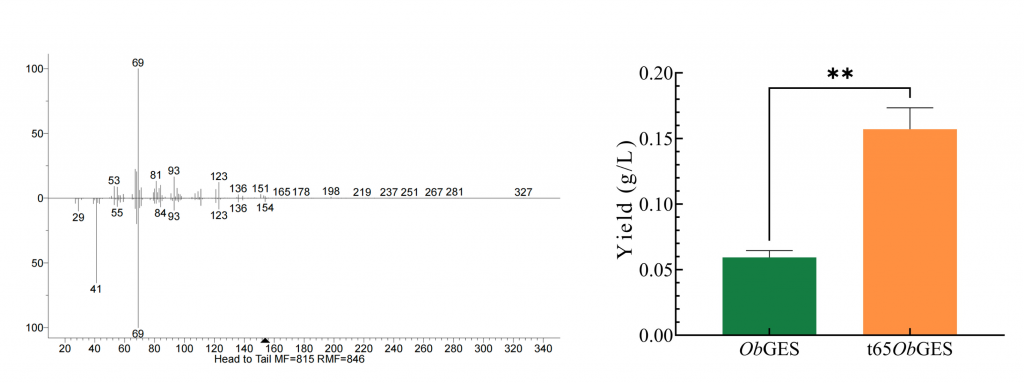
We identified several naturally occurring enzymes — such as chitinase, glucanase, and lysozyme — that can inhibit and kill mold. In addition, we incorporated a monoterpenoid called geraniol, which not only suppresses mold growth but also gives out a pleasant rose-like scent. To address geraniol’s high volatility and poor solubility, we designed a γ-cyclodextrin-geraniol inclusion complex, using encapsulation to enhance its stability and water-solubility [see lower right image].
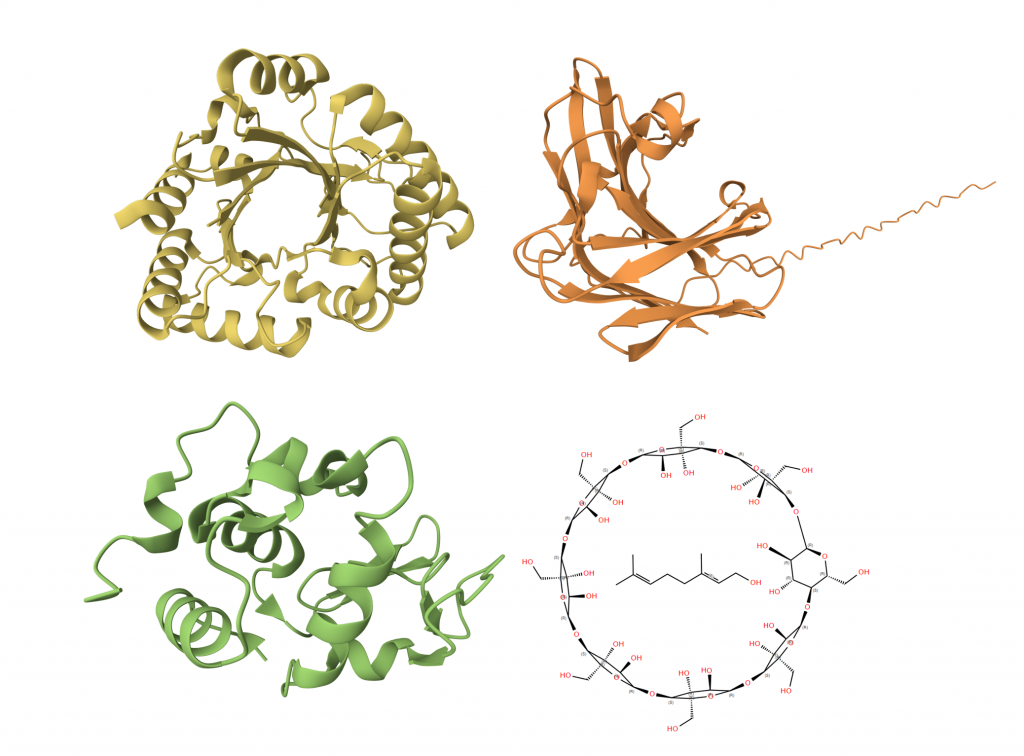
The cell wall of mold is primarily composed of chitin and glucan. Therefore, enzymes that break down these substances are highly effective at killing mold. Lysozyme, carrying a positive charge, interacts with the negatively charged mold cell wall, leading to its disruption.
To express these enzymes, we utilized E. coli and yeast as chassis organisms. Gene fragments encoding the enzymes were inserted into these microbes, enabling them to produce the target enzyme proteins. As for geraniol, we integrated the MVA pathway alongside with the geraniol synthesis pathway into engineered E. coli for heterologous synthesis.
However, the initial experiments were challenging. Some enzymes failed to express, others showed no activity at all, and some had only minimal hydrolytic activity. Through persistent investigation, we learned that modifying the protein sequences of these enzymes could enhance their various characteristics.

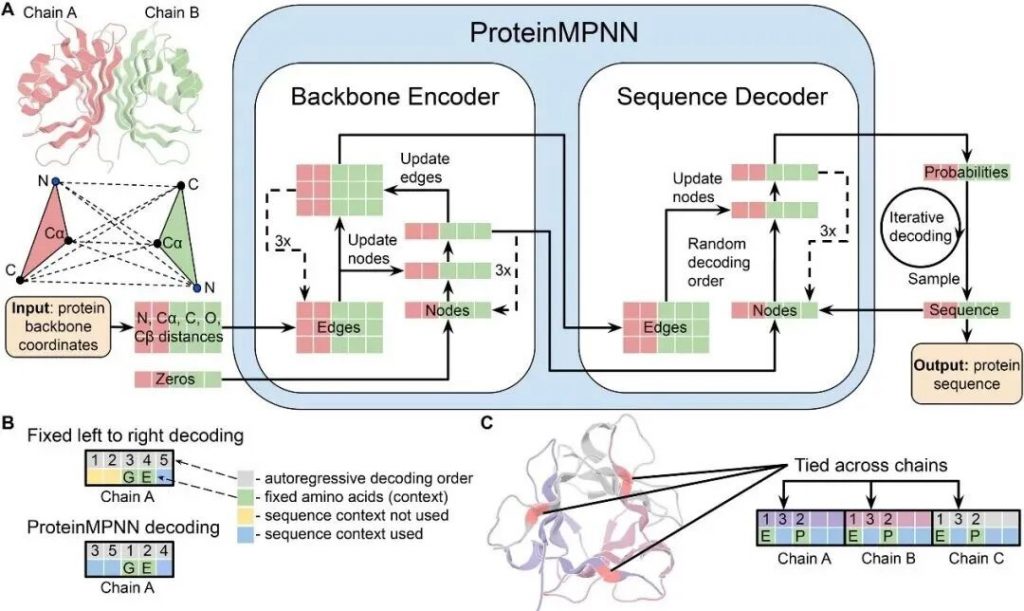
We employed the AI modelling tool MPNN (Message Passing Neural Networks). By inputting the enzyme protein structures, MPNN generates sequences that are not only structurally compatible but also predicted to exhibit enhanced stability, improved folding efficiency, and stronger interactions with ligands.
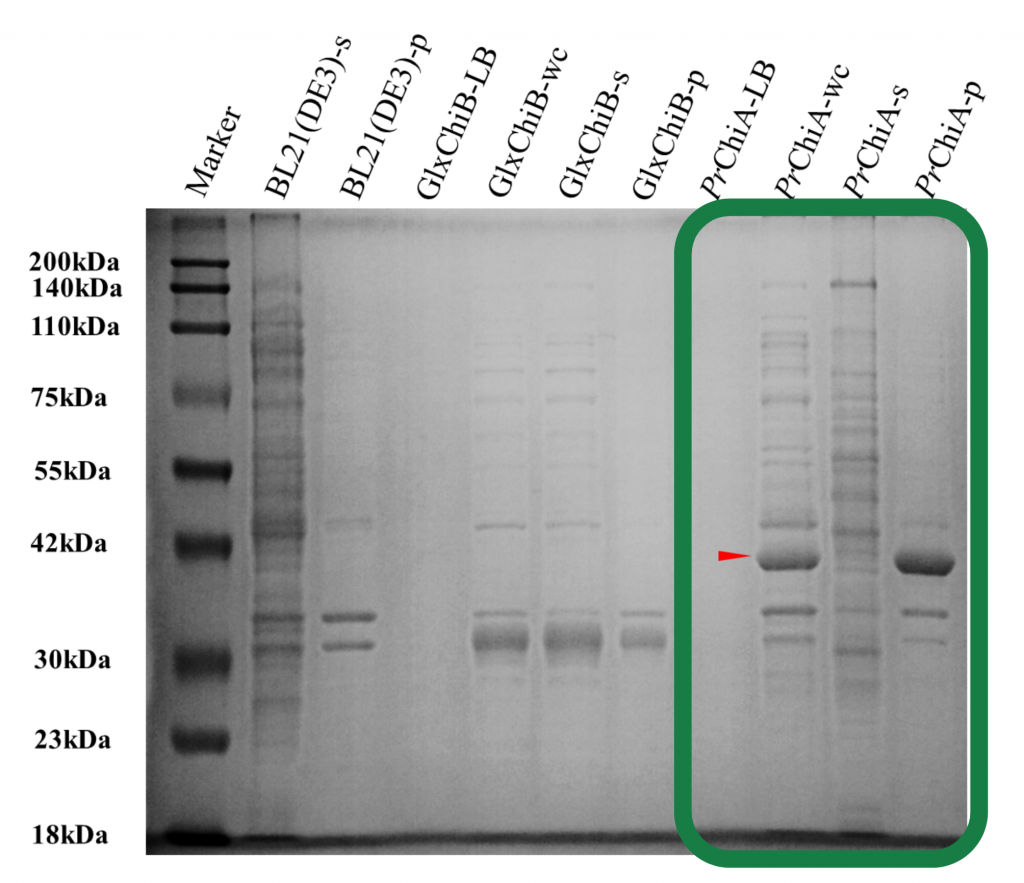
Guided by MPNN, one of our key enzymes, PrChiA, finally achieved successful soluble expression with detectable activity. That night, our modeling advisor, Andy, stayed up throughout the entire night.
Furthermore, through interviews with various experts and potential users, we identified air conditioners as a major hotspot for mold accumulation. To address this, we designed a CBM fusion protein by incorporating an additional carbohydrate-binding module into our enzymes. This design was proven to be highly successful—the binding module not only adhered strongly to various surfaces but also enhanced our enzymes’ catalytic activity.
The terpene production also achieved significant success. By truncating the signal peptide, we increased the geraniol yield by nearly two-fold. GC-MS results also indicated remarkably high purity.
Beyond our experimental design, we also conducted extensive human practices and educational initiatives. These efforts were aimed at deepening the understanding of our project’s context and communicating the appeal of synthetic biology to the broader public. iGEM recognized these contributions by awarding our team both the Best Integrated Human Practices and Best Education prizes.
During the process, we embarked on several trips for interviews. We traveled to the University of Guangzhou to meet a professor specializing in bio-enzymes, visited Gree Electric in Zhuhai to learn about air conditioner designs, and went to the Guangzhou Institute of Microbiology to consult two experts in mold remediation… Even on those typhoon days, we continued our trip, traveling by car, aiming to obtain the most comprehensive and detailed analyses and insights from these specialists.


That laboratory in that early March was where our dreams began. The following eight months of relentless effort have given us far more than just honors.
We transformed from students who only knew how to solve textbook problems, into beginners in molecular biology, skilled in using gel electrophoresis apparatus, SnapGene, and Maestro. We moved from presenting in front of classroom projectors, to the world stage of the Grand Jamboree; from End of Semester exam rooms, to a massive presentation hall filled with over 400 judges.
The bonds between our team members evolved from strangers eight months ago, to the closest friends with shared interest and passion. We grew from lazy chaps who used to sleep until 11 a.m., into dedicated workaholics, working 18 hours a day, and doing experiments in the lab until 3 a.m.
iGEM has bestowed us with capability, confidence, friendship, passion, joy… A single competition, an unforgettable journey. While the competition has concluded, the journey continues. The story of GreatBay-SCIE is far from over. Next year’s stage will still be ours!
References:
Dauparas, J., et al. “Robust Deep Learning–based Protein Sequence Design Using ProteinMPNN.” Science, vol. 378, no. 6615, Sept. 2022, pp. 49–56, doi:10.1126/science.add2187.
visit our wiki at https://2025.igem.wiki/greatbay-scie/index.html
- Article / Tom Lu


















